Abstract
Definition of MHC class I ligands of rhesus macaque killer cell Ig-like receptors (KIRs) is fundamental to NK cell biology in this species as an animal model for infectious diseases, reproductive biology, and transplantation. To provide a more complete foundation for studying NK cell responses, rhesus macaque KIRs representing common allotypes of lineage II KIR genes were tested for interactions with MHC class I molecules representing diverse Macaca mulatta (Mamu)-A, -B, -E, -F, -I, and -AG alleles. KIR–MHC class I interactions were identified by coincubating reporter cell lines bearing chimeric KIR-CD3ζ receptors with target cells expressing individual MHC class I molecules and were corroborated by staining with KIR IgG-Fc fusion proteins. Ligands for 12 KIRs of previously unknown specificity were identified that fell into three general categories: interactions with multiple Mamu-Bw4 molecules, interactions with Mamu-A–related molecules, including allotypes of Mamu-AG and the hybrid Mamu-B*045:03 molecule, or interactions with Mamu-A1*012:01. Whereas most KIRs found to interact with Mamu-Bw4 are inhibitory, most of the KIRs that interact with Mamu-AG are activating. The KIRs that recognize Mamu-A1*012:01 belong to a phylogenetically distinct group of macaque KIRs with a 3-aa deletion in the D0 domain that is also present in human KIR3DL1/S1 and KIR3DL2. This study more than doubles the number of rhesus macaque KIRs with defined MHC class I ligands and identifies interactions with Mamu-AG, -B*045, and -A1*012. These findings support overlapping, but nonredundant, patterns of ligand recognition that reflect extensive functional diversification of these receptors.
Introduction
Natural killer cell responses in humans and other primate species are regulated in part by interactions between two highly polymorphic sets of molecules: the killer cell Ig-like receptors (KIRs) expressed on the surface of NK cells and their MHC class I ligands on target cells (1, 2). As indicated by their nomenclature, KIRs typically have two or three extracellular Ig-like domains (2D or 3D) and either long (L) or short (S) cytoplasmic tails. KIRs with long cytoplasmic tails contain a pair of ITIMs that transduce inhibitory signals, whereas those with short cytoplasmic tails associate with adaptor molecules such as DAP12 or FcεRIγ to transduce activating signals (2, 3). Inhibitory KIRs normally suppress NK cell responses through interactions with MHC class I ligands on the surface of heathy cells. However, if these interactions are somehow perturbed, for instance as a result of MHC class I downregulation in virus-infected cells or deletion of MHC class I genes in tumor cells, the loss of these inhibitory signals can trigger an NK cell response (4–9). Although the molecular mechanisms are not as well defined, activating KIRs can also interact with MHC class I molecules on unhealthy cells to directly trigger an NK cell response (2, 10–12).
Polymorphisms in the KIR and HLA class I genes are associated with differences in the course of infection for a number of human viral pathogens and with the outcome of cancer immunotherapy (12–19). KIR and HLA class I polymorphisms have also been linked to complications during pregnancy and to the success of hematopoietic stem cell and solid organ transplantation (20–23). However, studies to address the immunological mechanisms underlying these observations have been hampered by the lack of a suitable animal model. Mice and other rodents do not have KIRs, but instead express C type lectin-like molecules on their NK cells as polymorphic receptors for MHC class I ligands (2, 24). Moreover, due to the rapid pace of KIR and MHC class I evolution (25–27), it is not possible to predict KIR ligands in nonhuman primates based on sequence similarity with human KIRs.
In contrast to humans, which have three classical HLA class I genes (HLA-A, -B, and -C), Old World monkeys express an expanded array of polymorphic MHC class I genes related to HLA-A and -B. However, these species do not have an HLA-C ortholog, as MHC-C arose in hominids as a duplication of an MHC-B gene after their divergence from Old World monkeys (28, 29). The MHC class I haplotypes of Old World monkeys are polygenic, and rhesus macaques typically express 2 or 3 Macaca mulatta (Mamu)-A genes and 4–11 Mamu-B genes (30–32). Depending on their transcriptional abundance, the MHC class I genes of macaques can be further classified as “major” or “minor” loci (32). Products of the major genes exhibit the greatest polymorphism and present peptides to virus-specific CD8+ T cells (33), suggesting functional similarities to HLA-A and -B in humans.
Four nonclassical MHC class I genes have also been identified in Old World monkeys. Orthologs of HLA-E and -F (Mamu-E and -F in rhesus macaques) are well conserved and serve similar functions as their human counterparts (34–37). Macaques and other Old World monkeys also have an ortholog of HLA-G; however the MHC-G genes of these species have accumulated multiple stop codons and no longer encode functional proteins (38, 39). In the absence of a functional MHC-G gene, Old World monkeys have evolved duplicated MHC-A genes, termed MHC-AG (Mamu-AG in rhesus macaques), that appear to serve a similar function (40–43). Similar to HLA-G, Mamu-AG has limited polymorphism, a truncated cytoplasmic tail, and is only expressed on placental trophoblasts (40, 41, 44). Mamu-AG is therefore thought to contribute to pregnancy success through interactions with receptors on NK cells and myeloid cells that promote placental vascularization, facilitate fetal tolerance, and protect the maternal–fetal interface from invading microorganisms (44–46). Rhesus macaques also express Mamu-I, which is the product of a fixed duplication of an MHC-B gene with limited sequence variation (47).
In concert with the tandem duplication of their Mamu-A and -B genes, macaques have evolved an expanded complement of lineage II KIRs that encode 3D KIRs for Mamu-A and -B ligands (26, 48–52). Whereas humans only have two lineage II KIR genes that encode KIR3DL1/S1 and KIR3DL2 as receptors for HLA-Bw4 and -A3/A11, respectively, rhesus macaques have at least 19 lineage II KIRs that encode inhibitory and activating receptors for Mamu-A and -B molecules (50, 52–56). Furthermore, in contrast to humans, which have multiple lineage III KIR genes encoding KIR2DL/S receptors for HLA-C1 and -C2 ligands (1, 2), macaques only have two lineage III KIR genes: one is a pseudogene (KIR3DP1) and the other (KIR1D) expresses a single-domain KIR of uncertain function (52). Thus, whereas human NK cell regulation predominantly depends on lineage III KIR interactions with HLA-C ligands, macaque NK cells are dependent on lineage II KIR interactions with Mamu-A and -B ligands (52).
The rhesus macaque has become an increasingly valuable animal model for infectious diseases (57–67 and S. Lu, Y. Zhao, W. Yu, Y. Yang, J. Gao, J. Wang, D. Kuang, M. Yang, J. Yang, C. Ma, et al., manuscript posted on bioRxiv, DOI: 10.1101/2020.04.08.031807), reproductive biology (44, 68), and transplantation (69, 70). Although MHC class I ligands have been identified for a few macaque KIRs (71–75), ligands for most of these receptors remain undefined. We therefore performed a systematic survey of MHC class I interactions for common lineage II KIR allotypes representing 16 predicted genes of Indian-origin rhesus macaques. By measuring the responses of KIR-CD3ζ–transduced reporter cell lines to target cells expressing individual MHC class I molecules and confirming interactions by staining with KIR IgG-Fc domain (KIR-Fc) fusion proteins, we identified MHC class I ligands for 12 rhesus macaque KIRs of previously undefined specificity. This represents the most comprehensive analysis of ligand recognition by macaque KIRs to date and reveals novel interactions as well as extensive functional diversification of these receptors.
Materials and Methods
Jurkat NFAT luciferase reporter cells expressing chimeric KIR-CD3ζ receptors
Common alleles representing 16 predicted lineage II KIR genes of Indian-origin rhesus macaques were selected from the Immuno Polymorphism Database (https://www.ebi.ac.uk/ipd/) and from KIR genotyping data available from animals at the Wisconsin National Primate Research Center. cDNA sequences encoding the leader peptide of Mamu-KIR3DL05*008 followed by a Flag-tag (DYKDDDDK) and the D0, D1, D2, and stem domains of each of the rhesus macaque KIR alleles listed below were synthesized (Integrated DNA Technologies) and cloned into pQCXIP in-frame with sequences encoding the transmembrane and cytoplasmic domains of human CD3ζ. All of the KIR-CD3ζ constructs therefore encode the same leader peptide, Flag-tag, and CD3ζ domains, but different KIR extracellular domains. Accuracy of all plasmid constructs was verified by Sanger sequencing of the entire open reading frame. Retroviral vectors were packaged into vesicular stomatitis virus (VSV)-G–pseudotyped murine leukemia virus particles by cotransfecting GP2-293 cells (Clontech Laboratories) with pQCXIP constructs and pVSV-G (Clontech Laboratories). Jurkat NFAT luciferase (JNL) cells (Signosis) were transduced via spinoculation for 1 h with filtered supernatant collected from transfected GP2-293 cells and 2 d later were placed under selection in RPMI 1640 medium supplemented with 10% FBS, l-glutamine, penicillin, and streptomycin (R10 medium) plus 0.1 mg/ml hygromycin (Invitrogen) to maintain the luciferase reporter gene. Selection for the KIR-CD3ζ receptor was performed by increasing the puromycin (Invitrogen) concentration to 1.0 µg/ml during 3–4 wk and then maintaining at 0.5 µg/ml. The GenBank (https://www.ncbi.nlm.nih.gov/genbank/) accession numbers for rhesus macaque KIR and human CD3ζ sequences are as follows: Mamu-KIR3DL02*004:01 (LT963635.1), -KIR3DLw03*002 (EU419055.1), -KIR3DL04*001:02 (GU299490.1), -KIR3DL07*004 (EU419060.1), -KIR3DL10*002:01 (GU112259.1), -KIR3DL11*012 (LS997637.1), -KIR3DLw34*002 (previously -KIR3DLw03*003; EU419031.1), -KIR3DS01*003 (GU564161.1), -KIR3DS02*004:02 (GU014296.1), -KIR3DS03*003 (EU702454.1), -KIR3DS04*003:01 (EU419029.1), -KIR3DS05*002:01 (GU112262.1), -KIR3DS06*002:02 (GU112298.1), -KIR3DSw07*001 (GU112272.1), -KIR3DSw08*001 (AY505479.1), -KIR3DSw09*003 (MF164924.1) and CD3ζ (J04132.1).
Rhesus macaque MHC class I–expressing 721.221 cells
MHC class I sequences were selected from the Immuno Polymorphism Database (https://www.ebi.ac.uk/ipd/) and from previous studies to represent phylogenetically distinct Mamu-A, -B, -E, -F, -I, and -AG alleles of unique genes based on segregation analyses and haplotype sequencing (30–32, 72, 74, 75). Codon-optimized cDNA sequences encoding rhesus macaque MHC class I H chain constructs were synthesized (Integrated DNA Technologies) and cloned into pQCXIP or pQCXIN and verified by Sanger sequencing of the entire open reading frame. Retroviral vectors were packaged into VSV-G–pseudotyped murine leukemia virus particles as described above. 721.221 cells were transduced via spinoculation for 1 h with filtered supernatant collected from transfected GP2-293 cells and placed under selection in R10 medium containing 0.4 µg/ml puromycin or 0.5 mg/ml G418, depending on the vector, to select for MHC class I expression. Selection for the MHC class I receptor was performed for pQCXIN constructs by increasing the G418 (Enzo Life Sciences) concentration to 1.0 mg/ml (based on active units) and then maintaining at 0.5 mg/ml and for pQCXIP constructs by increasing the puromycin (Invitrogen) concentration to 1.0 µg/ml and then maintaining at 0.4 µg/ml. The GenBank (https://www.ncbi.nlm.nih.gov/genbank/) accession numbers for all of the rhesus macaque MHC class I alleles used in this study are as follows: Mamu-A1*001:01:01:01 (LT908117), -A1*002:01:01:01 (LN899621), -A1*004:01:01:01 (FN396407.1), -A1*008:01:01:01 (LN899628), -A1*011:01:01:01 (AJ542579), -A1*012:01:01:01 (AF157398.1), -A2*05:02:01:01 (LN899639), -A2*05:04:01:01 (LT908121.1), -A3*13:02:01:01 (LN899654.1), -A3*13:03:01:01 (AF157401), -A4*14:03:01:01 (GU080236.1), -A6*01:03:02:01 (EF602318.1), -B*001:01:02:01 (LM608018), -B*002:01:01:01 (LN851852), -B*005:01:01:01 (LM608023), -B*007:01:01:01 (U41829.1), -B*008:01:01:01 (U41830), -B*015:01 (AM902541.1), -B*017:01:01:01 (AF199358), -B*022:01:01:01 (LN899675), -B*036:01:01:01 (AJ556886.1), -B*041:01:01:01 (LN899682), -B*043:01:01:01 (AJ556893), -B*045:03:01:01 (LM608047), -B*065:01:01:01 (AJ620416.1), -I*01:01:01 (LT908864), -E*02:01:02 (previously E*02:01; NM_001114966.1) -F*01:01 (LT899414), -AG1*05:01 (FJ409466), -AG2*01:01 (previously AG*01:01; U84783.1), AG2*01:02:01:01 (LR989653)-AG3*02:01:01:01 (U84785.1), -AG3*03:01:01:01 (U84787.1), -AG4*08:01:01:01 (LR989670), -AG4*09:02:02:02 (LR990451), -AG5*07:10:01:02 (LR989642), and -AG6*04:01:01:01 (LR989697). Only a partial-length cDNA sequence was available for -AG1*05:01. For the construct expressing this allele, sequences encoding the α3-, transmembrane, and cytoplasmic domains were inferred from the Mamu-AG consensus sequence.
KIR-Fc fusion proteins
All KIR-Fc fusion constructs were cloned into an in-house–generated expression construct (pCIW) that contains the following regulatory elements: CMV promoter/enhancer, SV40 intron, woodchuck hepatitis virus posttranscriptional regulatory element, and SV40 polyadenylation signal. The constructs encoded an artificial “secrecon” leader peptide (76) followed by KIR ectodomain fused to the Fc domain of murine IgG2A. Following cloning of the initial KIR3DL01 construct using a synthetic gBlock DNA fragment (Integrated DNA Technologies), all other KIR fragments were cloned in place of the KIR3DL01 using the QuickChange cloning protocol as described by Klenchin et al. (77). All fusion proteins were expressed in Expi-293F cells in 250–280 ml culture using the ExpiFectamine 293 transfection kit (Life Technologies) according to the manufacturer’s recommendations. Five to 6 d after transfection, the supernatant was collected, filtered through a 0.45-µm polyethersulfone filter, and loaded on a rProtein A GraviTrap column (GE Healthcare). After thorough washing of the column with PBS (at least 30 ml), KIR-Fc fusion proteins were eluted with 3 ml of Gentle Ag/Ab buffer (Pierce) and buffer exchanged into 50 mM sodium citrate (pH 6.5) using three cycles of concentration/dilution in an Amicon Ultra-15 50K centrifugal filter unit (Millipore). The final concentrations of the purified KIR-Fc fusion proteins were calculated from absorbance at 280 nm using theoretical extinction coefficients calculated by ProtParam (https://web.expasy.org/protparam/). The purified protein preparations were clarified by centrifugation at 30,000 × g for 30 min at 23°C, sodium azide was added to a final concentration of 3 mM, and the supernatants were snap-frozen in liquid nitrogen in 50-µl aliquots and stored at −80°C.
Flow cytometry
KIR-CD3ζ expression on the surface of JNL cells and MHC class I expression on 721.221 cells was confirmed by staining with near-infrared (IR) fluorescent dye (Invitrogen), washed twice with FACS buffer, followed by 30 min staining at room temperature with PE-conjugated anti-Flag (clone REA216, Miltenyi Biotec) or pan-HLA class I–specific Abs (clone W6/32, Life Technologies) and fixed in 2% formaldehyde in PBS. To assess KIR binding, MHC class I–transduced 721.221 cells were stained for 30 min at room temperature with near-IR fluorescent dye, washed twice with FACS buffer, and stained for 30 min at room temperature with KIR-Fc fusions proteins (10–20 µg/ml). The cells were then washed twice with FACS buffer, stained for 45 min with Alexa Fluor 647–conjugated goat anti-mouse IgG (RRID AB_2338925, Jackson ImmunoResearch Laboratories), washed twice, and stained for 30 min at room temperature with the PE-conjugated pan-MHC class I–specific Ab W6/32. The cells were washed again and fixed in 2% formaldehyde in PBS. All flow cytometry data were collected using a FACSymphony A3 Cell Analyzer (BD Biosciences) and analyzed using FlowJo version 10.8.0. After gating on viable cells, KIR-CD3ζ (Flag-tag) and MHC class I staining was compared with parental JNL and 721.221 cells, respectively. KIR-MHC class I binding was similarly assessed by comparing KIR-Fc versus W6/32 staining on the surface of MHC class I–transduced 721.221 cells to their levels of nonspecific staining on parental 721.221 cells.
KIR-CD3ζ JNL reporter assays
KIR-CD3ζ JNL cells and MHC class I–transduced 721.221 cells were cocultured at a 1:1 ratio (1 × 105 cells each) overnight at 37°C and 5% CO2 in R10 medium without antibiotics. KIR-CD3ζ JNL cells were also incubated with parental 721.221 cells as a negative control and with anti–Flag-tag (5 µg/ml) (clone 5A8E5, GenScript) and goat anti-mouse (10 µg/ml) (Poly4053, BioLegend) Abs as a positive “X-link” control. Each combination was tested in triplicate wells (100 µl/well). After 12–18 h, britelite Plus luciferase substrate (PerkinElmer) was added to each well (100 µl/well) and the relative light units (RLU) of luciferase activity were measured using a VICTOR X4 multiplate reader (PerkinElmer).
Alignments and phylogenetic trees
Nucleic acid sequences of KIR and MHC class I molecules were obtained from the Immuno Polymorphism Database (https://www.ebi.ac.uk/ipd/) and aligned using Geneious Prime 2021.2.2 (Biomatters). Phylogenetic trees were constructed using raw difference neighbor-joining analysis of the α1–α3 domains of Mamu-MHC and the D0–D2 domains of human and rhesus macaque KIRs employing the MAFFT server, version 7 (https://mafft.cbrc.jp/alignment/server/) (78, 79). All analyses used G-INS-i iterative refinement with 1000 bootstrap replicates. Analysis parameters included scoring matrix of 200PAM/k = 2, gap penalty 1.53, offset value of 0, and nzero alignment scoring. Trees were viewed using Phylo.io version 1.0.0 (80). For the analysis of KIR sequences, human lineage II KIRs were included as an outgroup (KIR3DL1*001, AY760023.1 and KIR3DL2*001, MN196233.1).
Statistical analysis
The luciferase induction by KIR-CD3ζ JNL cells (RLU data) was analyzed by one-way ANOVA for multiple comparisons of mean responses to each of the MHC class I–expressing 721.221 cells with mean responses to the parental 721.221 cells. Statistical analyses were performed using GraphPad Prism for Mac OS, version 10.8.0.
Results
Extensive sequencing and segregation analyses of KIR transcripts support the existence of at least 22 KIR genes in Indian-origin rhesus macaques (53–56). Although our group and others have identified ligands for a few rhesus macaque KIRs (71–75), ligands for most of the KIRs in this species remain undefined. To provide a more complete foundation for studying NK cell biology in this species, we investigated the MHC class I interactions of 16 KIRs representing common alleles of distinct lineage II genes in animals of Indian descent (Fig. 1A). Jurkat cells harboring an NFAT-inducible luciferase reporter gene (JNL cells) were transduced with retroviral vectors expressing chimeric KIR-CD3ζ receptors consisting of the extracellular domains of macaque KIRs (D0, D1, D2, and stem domains) fused to the transmembrane and cytoplasmic domains of human CD3ζ. To confirm surface expression, a Flag-tag was appended to the N terminus of the D0 domain of each KIR-CD3ζ receptor (Fig. 1B). 721.221 cells were similarly transduced with vectors expressing rhesus macaque MHC class I molecules, and surface expression was confirmed with a pan-MHC class I Ab (Supplemental Figs. 1–3). After coincubating the KIR-CD3ζ JNL cells overnight with MHC class I–transduced 721.221 cells, ligand recognition was detected by the MHC class I–dependent upregulation of luciferase. KIR–MHC class I interactions were also corroborated by staining MHC class I–expressing 721.221 cells with KIR-Fc fusion proteins consisting of the extracellular domains of macaque KIRs fused to the Fc domain of murine IgG2A.
Rhesus macaque KIRs and KIR-CD3ζ expression on JNL cells. (A) Alignment of amino acid sequences for the D0, D1, and D2 domains of rhesus macaque KIRs. Shaded sequences correspond to predicted MHC class I contact sites based on the crystal structure of HLA-B*57 in complex with human KIR3DL1 (89). Positions of identity are indicated by periods, and amino acid differences are identified by their single-letter code. (B) KIR-CD3ζ–transduced JNL cells were stained with anti–Flag-tag Ab and near-IR Live/Dead stain. After excluding dead cells, the fluorescence intensity of KIR-CD3ζ (Flag-tag) staining on KIR-CD3ζ–transduced JNL cells (open) was compared with background staining on parental JNL cells (shaded).
Rhesus macaque KIRs and KIR-CD3ζ expression on JNL cells. (A) Alignment of amino acid sequences for the D0, D1, and D2 domains of rhesus macaque KIRs. Shaded sequences correspond to predicted MHC class I contact sites based on the crystal structure of HLA-B*57 in complex with human KIR3DL1 (89). Positions of identity are indicated by periods, and amino acid differences are identified by their single-letter code. (B) KIR-CD3ζ–transduced JNL cells were stained with anti–Flag-tag Ab and near-IR Live/Dead stain. After excluding dead cells, the fluorescence intensity of KIR-CD3ζ (Flag-tag) staining on KIR-CD3ζ–transduced JNL cells (open) was compared with background staining on parental JNL cells (shaded).
Mamu-Bw4 recognition by multiple rhesus macaque KIRs
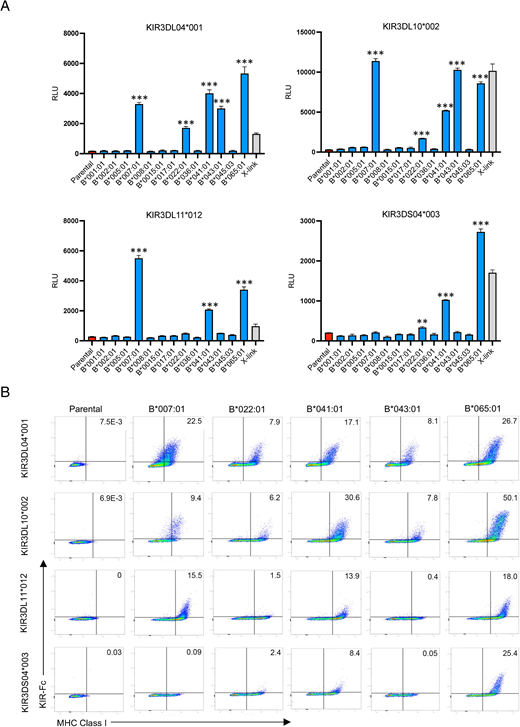
Four of the rhesus macaque KIRs recognized Mamu-Bw4 ligands. KIR3DL04*001- and KIR3DL10*002-CD3ζ JNL cells responded to 721.221 cells expressing Mamu-B*007:01, -B*022:01, -B*041:01, -B*043:01, and -B*065:01 (Fig. 2A, Supplemental Fig. 4A). Similarly, KIR3DL11*012-CD3ζ JNL cells responded to Mamu-B*007:01, -B*041:01, and -B*065:01, and KIR3DS04*003-CD3ζ JNL cells responded to Mamu-B*22:01, -B*041:01, and -B*065:01 (Fig. 2A, Supplemental Fig. 4A). These interactions were corroborated by KIR-Fc staining of 721.221 cells expressing these molecules (Fig. 2B, Supplemental Fig. 4B).
KIR3D04*001, KIR3DL10*002, KIR3DL11*012, and KIR3DS04*003 interact with multiple Mamu-Bw4 ligands. (A) KIR3DL04*001-, KIR3DL10*002-, KIR3DL11*012-, and KIR3DS04*003-CD3ζ JNL cells were incubated with 721.221 cells expressing the indicated Mamu-B molecules. The bar graphs represent the mean and SD (error bars) of luciferase activity (RLU) from triplicate wells of KIR-CD3ζ JNL cells incubated with Mamu-B+721.221 cells (blue), parental 721.221 cells (red), and anti–Flag-tag plus anti-mouse Abs (X-link, gray). **p = 0.0002, ***p < 0.0001, by one-way ANOVA with a Dunnett test. These results are representative of at least three independent experiments. (B) Parental and Mamu-B+721.221 cells were stained with near-IR Live/Dead dye, KIR3DL04*001-, KIR3DL10*002-, KIR3DL11*012-, and KIR3DS04*003-Fc, followed by goat anti-mouse IgG and MHC class I–specific Ab (W6/32).
KIR3D04*001, KIR3DL10*002, KIR3DL11*012, and KIR3DS04*003 interact with multiple Mamu-Bw4 ligands. (A) KIR3DL04*001-, KIR3DL10*002-, KIR3DL11*012-, and KIR3DS04*003-CD3ζ JNL cells were incubated with 721.221 cells expressing the indicated Mamu-B molecules. The bar graphs represent the mean and SD (error bars) of luciferase activity (RLU) from triplicate wells of KIR-CD3ζ JNL cells incubated with Mamu-B+721.221 cells (blue), parental 721.221 cells (red), and anti–Flag-tag plus anti-mouse Abs (X-link, gray). **p = 0.0002, ***p < 0.0001, by one-way ANOVA with a Dunnett test. These results are representative of at least three independent experiments. (B) Parental and Mamu-B+721.221 cells were stained with near-IR Live/Dead dye, KIR3DL04*001-, KIR3DL10*002-, KIR3DL11*012-, and KIR3DS04*003-Fc, followed by goat anti-mouse IgG and MHC class I–specific Ab (W6/32).
Mamu-B*007:01, -B*022:01, -B*041:01, -B*043:01, and -B*065:01 share a Bw4 motif at residues 77–83 (Supplemental Fig. 2), which was previously shown to contribute to recognition by KIR3DL01 as well as three other rhesus macaque KIRs (72, 74). The identification of Mamu-Bw4 ligands for KIR3DL04*001, KIR3DL10*002, KIR3DL11*012, and KIR3DS04*003 doubles the number of rhesus macaque KIRs known to interact with this group of molecules and reveals an extraordinary expansion of the lineage II KIRs in macaques with Bw4 specificity (Table I). Differences in the particular Mamu-Bw4 molecules recognized by these KIRs and the relative strength of these interactions suggest extensive functional diversification of these receptors.
| KIR Genea . | Allotypeb . | MHC Class I Ligandsc . | Groupd . | Referencese . |
|---|---|---|---|---|
| KIR3DL01 | KIR3DL01*012 | Mamu-B*007:01, -B*022:01, -B*041:01, -B*043:01, -B*065:01 | Bw4 | (72) |
| KIR3DL02 | KIR3DL02*004 | Mamu-A1*012:01 | A | This study |
| KIR3DLw03 | KIR3DLw03*002 | Mamu-A1*012:01 | A | This study |
| KIR3DL04 | KIR3DL04*001 | Mamu-B*007:01, -B*022:01, -B*041:01, -B*043:01, -B*065:01 | Bw4 | This study |
| KIR3DL05 | KIR3DL05*008 | Mamu-A1*002:01, -A3*13:03, -B*036:01, -AG2*01:01, -AG1*05:01 | AG/A-related | (71, 73, 74) |
| KIR3DL06 | KIR3DL06*001 | Mamu-B*007:01, -B*022:01, -B*041:01, -B*043:01, -B*065:01 | Bw4 | (74) |
| KIR3DL07 | KIR3DL07*004 | Mamu-AG2*01:01, -AG2*01:02, -AG6*04:01, -B*045:03 | AG/A-related | This study |
| KIR3DL08 | KIR3DL08*002 | Mamu-B*002:01, -B*007:01, -B*022:01, -B*041:01, -B*065:01 | Bw4 | (74) |
| KIR3DL10 | KIR3DL10*002 | Mamu-B*007:01, -B*022:01, -B*041:01, -B*043:01, -B*065:01 | Bw4 | This study |
| KIR3DL11 | KIR3DL11*012 | Mamu-B*007:01, -B*041:01, -B*065:01 | Bw4 | This study |
| KIR3DLw34 | KIR3DLw34*002 | None detectedf | This study | |
| KIR3DS01 | KIR3DS01*003 | Mamu-AG2*01:01, -AG6*04:01 | AG | This study |
| KIR3DS02 | KIR3DS02*004 | Mamu-AG2*01:01, -AG2*01:02, -AG6*04:01, -AG1*05:01, -B*045:03 | AG/A-related | This study, (75) |
| KIR3DS03 | KIR3DS03*003 | None detectedf | This study | |
| KIR3DS04 | KIR3DS04*003 | Mamu-B*022:01, -B*041:01, -B*065:01 | Bw4 | This study |
| KIR3DS05 | KIR3DS05*002 | None detectedf | This study | |
| KIR3DS06 | KIR3DS06*002 | Mamu-AG2*01:01, -AG1*05:01, -AG6*04:01 | AG | This study |
| KIR3DSw07 | KIR3DSw07*001 | Mamu-AG2*01:01, -AG6*04:01 | AG | This study |
| KIR3DSw08 | KIR3DSw08*001 | None detectedf | This study | |
| KIR3DSw09 | KIR3DSw09*003 | Mamu-AG2*01:01, -AG2*01:02, -AG1*05:01, -B*045:03 | AG/A-related | This study |
| KIR3DSw39 | KIR3DSw39*002 | Mamu-B*007:01, -B*022:01, -B*041:01, -B*043:01, -B*065:01 | Bw4 | (74) |
| KIR Genea . | Allotypeb . | MHC Class I Ligandsc . | Groupd . | Referencese . |
|---|---|---|---|---|
| KIR3DL01 | KIR3DL01*012 | Mamu-B*007:01, -B*022:01, -B*041:01, -B*043:01, -B*065:01 | Bw4 | (72) |
| KIR3DL02 | KIR3DL02*004 | Mamu-A1*012:01 | A | This study |
| KIR3DLw03 | KIR3DLw03*002 | Mamu-A1*012:01 | A | This study |
| KIR3DL04 | KIR3DL04*001 | Mamu-B*007:01, -B*022:01, -B*041:01, -B*043:01, -B*065:01 | Bw4 | This study |
| KIR3DL05 | KIR3DL05*008 | Mamu-A1*002:01, -A3*13:03, -B*036:01, -AG2*01:01, -AG1*05:01 | AG/A-related | (71, 73, 74) |
| KIR3DL06 | KIR3DL06*001 | Mamu-B*007:01, -B*022:01, -B*041:01, -B*043:01, -B*065:01 | Bw4 | (74) |
| KIR3DL07 | KIR3DL07*004 | Mamu-AG2*01:01, -AG2*01:02, -AG6*04:01, -B*045:03 | AG/A-related | This study |
| KIR3DL08 | KIR3DL08*002 | Mamu-B*002:01, -B*007:01, -B*022:01, -B*041:01, -B*065:01 | Bw4 | (74) |
| KIR3DL10 | KIR3DL10*002 | Mamu-B*007:01, -B*022:01, -B*041:01, -B*043:01, -B*065:01 | Bw4 | This study |
| KIR3DL11 | KIR3DL11*012 | Mamu-B*007:01, -B*041:01, -B*065:01 | Bw4 | This study |
| KIR3DLw34 | KIR3DLw34*002 | None detectedf | This study | |
| KIR3DS01 | KIR3DS01*003 | Mamu-AG2*01:01, -AG6*04:01 | AG | This study |
| KIR3DS02 | KIR3DS02*004 | Mamu-AG2*01:01, -AG2*01:02, -AG6*04:01, -AG1*05:01, -B*045:03 | AG/A-related | This study, (75) |
| KIR3DS03 | KIR3DS03*003 | None detectedf | This study | |
| KIR3DS04 | KIR3DS04*003 | Mamu-B*022:01, -B*041:01, -B*065:01 | Bw4 | This study |
| KIR3DS05 | KIR3DS05*002 | None detectedf | This study | |
| KIR3DS06 | KIR3DS06*002 | Mamu-AG2*01:01, -AG1*05:01, -AG6*04:01 | AG | This study |
| KIR3DSw07 | KIR3DSw07*001 | Mamu-AG2*01:01, -AG6*04:01 | AG | This study |
| KIR3DSw08 | KIR3DSw08*001 | None detectedf | This study | |
| KIR3DSw09 | KIR3DSw09*003 | Mamu-AG2*01:01, -AG2*01:02, -AG1*05:01, -B*045:03 | AG/A-related | This study |
| KIR3DSw39 | KIR3DSw39*002 | Mamu-B*007:01, -B*022:01, -B*041:01, -B*043:01, -B*065:01 | Bw4 | (74) |
Rhesus macaque lineage II KIR genes.
Representative KIR allotypes. KIR allotypes tested in this study are indicated in bold.
Novel MHC class I ligands defined in this study are indicated in bold.
Group of MHC class I ligands.
References for previously reported KIR ligands.
See Supplemental Fig. 8.
Multiple KIRs recognize Mamu-AG molecules
Six rhesus macaque KIRs were found to interact with Mamu-AG, a nonclassical MHC class I molecule expressed at the maternal–fetal interface of the placenta (40–44). JNL cells expressing KIR-CD3ζ chimeras for KIR3DL07*004, KIR3DS01*003, KIR3DS02*004, KIR3DS06*002, KIR3DSw07*001, and KIR3DSw09*003 all responded strongly to 721.221 cells expressing Mamu-AG2*01:01 and -AG6*04:01 (Fig. 3A, Supplemental Fig. 5A). In addition, KIR3DS02*004-CD3ζ JNL cells also engaged 721.221 cells expressing Mamu-AG2*01:02 (Fig. 3A). The binding of these KIRs to Mamu-AG2*01:01 and -AG6*04:01 was corroborated by KIR-Fc staining of Mamu-AG2–expressing 721.221 cells (Fig. 3B, Supplemental Fig. 5B). KIR-Fc staining also revealed detectable interactions with Mamu-AG1*05:01 for KIR3DS02*004, KIR3DS06*002, and KIR3DSw09*003 as well as Mamu-AG2*01:02 for KIR3DL07*004 and KIR3DSw09*003 that were not observed in cellular assays with KIR-CD3ζ JNL cells (Fig. 3B, Supplemental Fig. 5B). In this case, the detection of binding interactions with Mamu-AG1*05:01 and -AG2*01:02 that were not detectable with KIR-CD3ζ JNL cells may reflect greater sensitivity of KIR-Fc staining. It is possible that the dimeric nature of the KIR-Fc fusion proteins increases their avidity for MHC class I ligands via cooperative binding (81), making this assay more sensitive than the JNL reporter cell assay. It is also possible that the Flag-tag appended to the N terminus of the KIR-CD3ζ receptor may interfere with MHC class I interactions in some cases. These findings identify multiple Mamu-AG ligands for the products of six different KIR genes, five of which encode activating receptors. This brings the total number of rhesus macaque KIRs known to interact with Mamu-AG to seven (Table I).
KIR3DL07*004, KIR3DS01*003, KIR3DS02*004, KIR3DS06*002, KIR3DSw07*001, and KIR3DSw09*003 interact with Mamu-AG and -B*045:03. (A) KIR3DL07*004-, KIR3DS01*003-, KIR3DS02*004-, KIR3DS06*002-, KIR3DSw07*001-, and KIR3DSw09*003-CD3ζ JNL cells were incubated with 721.221 cells expressing the indicated Mamu-AG or -B molecules. The bar graphs represent the mean and SD (error bars) of luciferase activity (RLU) from triplicate wells of KIR-CD3ζ JNL cells incubated with Mamu-AG+721.221 cells (purple), Mamu-B+721.221 cells (blue), parental 721.221 cells (red), and anti–Flag-tag plus anti-mouse Abs (X-link, gray). **p < 0.005, ***p < 0.0001, by one-way ANOVA with a Dunnett test. These results are representative of at least three independent experiments. (B) Parental, Mamu-AG, and Mamu-B+721.221 cells were stained with near-IR Live/Dead dye, KIR3DL07*004-, KIR3DS01*003-, KIR3DS02*004-, KIR3DS06*002-, KIR3DSw07*001-, and KIR3DSw09*003-Fc, followed by goat anti-mouse IgG and MHC class I–specific Ab (W6/32).
KIR3DL07*004, KIR3DS01*003, KIR3DS02*004, KIR3DS06*002, KIR3DSw07*001, and KIR3DSw09*003 interact with Mamu-AG and -B*045:03. (A) KIR3DL07*004-, KIR3DS01*003-, KIR3DS02*004-, KIR3DS06*002-, KIR3DSw07*001-, and KIR3DSw09*003-CD3ζ JNL cells were incubated with 721.221 cells expressing the indicated Mamu-AG or -B molecules. The bar graphs represent the mean and SD (error bars) of luciferase activity (RLU) from triplicate wells of KIR-CD3ζ JNL cells incubated with Mamu-AG+721.221 cells (purple), Mamu-B+721.221 cells (blue), parental 721.221 cells (red), and anti–Flag-tag plus anti-mouse Abs (X-link, gray). **p < 0.005, ***p < 0.0001, by one-way ANOVA with a Dunnett test. These results are representative of at least three independent experiments. (B) Parental, Mamu-AG, and Mamu-B+721.221 cells were stained with near-IR Live/Dead dye, KIR3DL07*004-, KIR3DS01*003-, KIR3DS02*004-, KIR3DS06*002-, KIR3DSw07*001-, and KIR3DSw09*003-Fc, followed by goat anti-mouse IgG and MHC class I–specific Ab (W6/32).
Recognition of Mamu-B*045:03 by KIR3DL07*004, KIR3DS02*004, and KIR3DSw09*003
In addition to Mamu-AG, JNL cells expressing KIR3DL07*004-CD3ζ also responded to Mamu-B*045:03 (Fig. 3A, Supplemental Fig. 6A). KIR-Fc staining corroborated this interaction and additionally also revealed low but detectable interactions for Mamu-B*045:03 with KIR3DS02*004 and KIR3DSw09*003 (Fig. 3B, Supplemental Fig. 6B). To better understand how these KIRs, which predominately recognize Mamu-AG ligands, can also interact with B*045:03, we compared the amino acid sequences for Mamu-B*045:03 with other Mamu-A, -B, and -AG molecules. Although Mamu-B*045:03 resembles a typical Mamu-B molecule in its α2 domain, we noted a region of sequence similarity with other Mamu-A molecules in the α1 domain (Fig. 4A). These observations were also supported by phylogenetic analysis. Whereas the full-length nucleotide sequence for Mamu-B*045:03 clusters with Mamu-B molecules (Fig. 4B), the α1 domain is more similar to Mamu-A molecules (Fig. 4C). A similar pattern of clustering was observed for Mamu-B*036, which is a product of recombination between MHC-A and -B genes and was previously identified as a ligand for KIR3DL05 (74). Mamu-B*045:03 recognition by KIR3DL07*004, KIR3DS02*004, and KIR3DSw09*003 may therefore be explained by its origins as the product of recombination between ancestral MHC-A and -B genes.
Mamu-B*045:03 has an α1 domain similar to Mamu-A molecules. (A) Amino acid sequence alignment for the α1 and α2 domains of rhesus macaque MHC class I molecules. Residues 77–83 corresponding to the Bw4/Bw6 motif are underlined, and predicted KIR contact sites based on the crystal structure of HLA-B*57 in complex with human KIR3DL1 are shaded (89). Positions of identity are indicated by periods, and amino acid differences are identified by their single-letter code. (B and C) Phylogenetic trees of nucleotide sequence coding for the α1–α3 domains (B) or the α1 domain (C) of selected Mamu-A, -B, and -AG molecules. Neighbor-joining analysis was performed using the MAFFT server (version 7, https://mafft.cbrc.jp/alignment/server/). Bootstrap support of >50% is indicated.
Mamu-B*045:03 has an α1 domain similar to Mamu-A molecules. (A) Amino acid sequence alignment for the α1 and α2 domains of rhesus macaque MHC class I molecules. Residues 77–83 corresponding to the Bw4/Bw6 motif are underlined, and predicted KIR contact sites based on the crystal structure of HLA-B*57 in complex with human KIR3DL1 are shaded (89). Positions of identity are indicated by periods, and amino acid differences are identified by their single-letter code. (B and C) Phylogenetic trees of nucleotide sequence coding for the α1–α3 domains (B) or the α1 domain (C) of selected Mamu-A, -B, and -AG molecules. Neighbor-joining analysis was performed using the MAFFT server (version 7, https://mafft.cbrc.jp/alignment/server/). Bootstrap support of >50% is indicated.
Recognition of Mamu A1*012:01 by KIR3DLw03*002 and KIR3DL02*004
KIR3DLw03*002-CD3ζ JNL cells robustly responded to 721.221 cells expressing Mamu-A*012:01 (Fig. 5A, Supplemental Fig. 7A), but not to other MHC class I molecules tested in this study. The binding of KIR3DLw03*002 to Mamu-A*012:01 was corroborated by KIR-Fc staining (Fig. 5B, Supplemental Fig. 7B). An additional interaction between KIR3DL02*004 and Mamu-A1*012:01 was observed by KIR-Fc staining that was not detectable with KIR3DL02*004-CD3ζ JNL cells (Fig. 5, Supplemental Fig. 7). KIR3DLw03*002 and KIR3DL02*004 belong to a group of rhesus macaque KIRs with a characteristic deletion of 3 aa in the D0 domain (residues 37–39) (Fig. 1A).
KIR3DLw03*002 and KIR3DL02*004 interact with Mamu-A*012:01. (A) KIR3DLw03*002- and KIR3DL02*004-CD3ζ JNL cells were incubated with 721.221 cells expressing the indicated rhesus macaque MHC class I molecules. The bar graphs represent the mean and SD (error bars) of luciferase activity (RLU) from triplicate wells of KIR-CD3ζ JNL cells incubated with Mamu-A+721.221 (green), nonclassical (purple), and parental 721.221 cells (red) and anti–Flag-tag plus anti-mouse Abs (X-link, gray). ***p < 0.0001, by one-way ANOVA with a Dunnett test. The results are representative of at least three independent experiments. (B) Parental and Mamu-A+721.221 cells were stained with near-IR Live/Dead dye and KIR3DLw03*002- or KIR3DL02*004-Fc followed by goat anti-mouse IgG and MHC class I–specific Ab (W6/32).
KIR3DLw03*002 and KIR3DL02*004 interact with Mamu-A*012:01. (A) KIR3DLw03*002- and KIR3DL02*004-CD3ζ JNL cells were incubated with 721.221 cells expressing the indicated rhesus macaque MHC class I molecules. The bar graphs represent the mean and SD (error bars) of luciferase activity (RLU) from triplicate wells of KIR-CD3ζ JNL cells incubated with Mamu-A+721.221 (green), nonclassical (purple), and parental 721.221 cells (red) and anti–Flag-tag plus anti-mouse Abs (X-link, gray). ***p < 0.0001, by one-way ANOVA with a Dunnett test. The results are representative of at least three independent experiments. (B) Parental and Mamu-A+721.221 cells were stained with near-IR Live/Dead dye and KIR3DLw03*002- or KIR3DL02*004-Fc followed by goat anti-mouse IgG and MHC class I–specific Ab (W6/32).
Phylogenetic relationship of rhesus macaque KIRs is concordant with patterns of ligand recognition
To investigate the relationship between the extracellular domains of rhesus macaque KIRs and their patterns of ligand recognition, we constructed a phylogenic tree of the D0–D2 domains of rhesus macaque KIRs using human KIR3DL1 and KIR3DL2 as outgroups. The clustering of macaque KIRs generally reflected their ligand recognition. Six of the eight KIRs found to interact with Mamu-Bw4 ligands clustered together on a branch that includes KIR3DSw39*002, KIR3DL06*001, KIR3DS04*003, KIR3DL11*012, KIR3DL04*001, and KIR3DL08*002 (Fig. 6). Likewise, six of the seven KIRs found to interact with Mamu-AG ligands clustered together on a separate branch that includes KIR3DS06*002, KIR3DS01*003, KIR3DL07*004, KIR3DSw09*003, KIR3DS02*004, and KIR3DL05*008 (Fig. 6). KIR3DL10*002 and KIR3DSw07*001 were exceptions. Although KIR3DL10*002 and KIR3DSw07*001 interact with Mamu-Bw4 and -AG ligands, respectively, these receptors grouped together on the same branch (Fig. 6). KIR3DLw03*002 and KIR3DL02*004, which interact with Mamu-A1*012:01, were found on a distinct branch with two other KIRs for which ligands have not yet been identified (KIR3DS05*002 and KIR3DLw34*002) (Fig. 6). This group of KIRs is separated from the others by strong bootstrap support and is distinguished by the absence of 3 aa (residues 37–39) in the D0 domain (Fig. 1A) that are also missing in human KIR3DL1/S1 and KIR3DL2.
Phylogenetic clustering of rhesus macaque KIRs reflect their patterns of ligand recognition. Neighbor-joining analysis of nucleotide sequences coding for the D0–D2 domains of human and rhesus macaque KIRs was performed using the MAFFT server (version 7, https://mafft.cbrc.jp/alignment/server/). Bootstrap support of >50% is indicated. Human KIR3DL1 and KIR3DL2 were used included as an outgroup. Rhesus macaque KIRs are color coded according to their MHC class I ligands: unknown (gray), Mamu-Bw4 (green), Mamu-AG (dark blue), and Mamu-A1*012:01 (purple).
Phylogenetic clustering of rhesus macaque KIRs reflect their patterns of ligand recognition. Neighbor-joining analysis of nucleotide sequences coding for the D0–D2 domains of human and rhesus macaque KIRs was performed using the MAFFT server (version 7, https://mafft.cbrc.jp/alignment/server/). Bootstrap support of >50% is indicated. Human KIR3DL1 and KIR3DL2 were used included as an outgroup. Rhesus macaque KIRs are color coded according to their MHC class I ligands: unknown (gray), Mamu-Bw4 (green), Mamu-AG (dark blue), and Mamu-A1*012:01 (purple).
Discussion
Knowledge of the MHC class I ligands of rhesus macaques KIRs is fundamental to NK cell biology in this species as a preclinical model for infectious diseases, transplantation, and reproductive biology. Because of the rapid pace of KIR evolution in primates, it is not possible to predict the ligands for macaque KIRs on the basis of sequence similarity with their human counterparts. The KIR genes of macaques are evolving at a particularly rapid pace characterized by extensive gene duplication and recombination, generating allelic variation and haplotype diversity that greatly exceeds the genetic diversity of human KIRs (55, 56, 82). The rapid evolution of macaque lineage II KIRs appears to be a function of coevolution with the diversification of their MHC-A and -B genes (32, 52). Although the number of alleles and predicted KIR genes varies among geographically distinct populations (83), extensive sequencing and segregation analyses strongly support the existence of at least 22 distinct KIR genes in rhesus macaques of Indian origin (55, 56, 82). Of these, 19 are predicted to encode lineage II KIRs for Mamu-A and -B ligands. To provide a foundation for studying NK cell responses in macaques, we selected common allotypes representing 16 lineage II KIRs for analysis of their MHC class I interactions. By screening 721.221 cells expressing MHC class I molecules corresponding to 37 Mamu-A, -B, -E, -F, -I, and -AG alleles in assays with KIR-CD3ζ JNL reporter cell lines and by staining these MHC class I–expressing 721.221 cells with KIR-Fc fusion proteins, we identified ligands for 12 previously uncharacterized KIRs.
These analyses revealed three general patterns of ligand recognition: interactions with Mamu-Bw4 molecules, interactions with Mamu-A–related molecules, particularly Mamu-AG, or interactions with Mamu-A1*012:01. Of the 16 KIRs tested in this study, four (KIR3DL04*001, KIR3DL10*002, KIR3DL11*012, and KIR3DS04*003) were found to interact with molecules that contain a Bw4 motif at residues 77–83. This demonstrates a remarkable expansion of Bw4 specificity in the rhesus macaque. In contrast to humans, which only have a single KIR3DL1/S1 gene that encodes receptors for HLA-Bw4, rhesus macaques have at least eight lineage II KIRs that encode receptors for Bw4 molecules (Table I). Six of the KIRs in this study (KIR3DL07*004, KIR3DS01*003, KIR3DS02*004, KIR3DS06*002, KIR3DSw07*001, and KIR3DSw09*003) were found to interact with Mamu-AG2*01:01 and AG6*04:01, either alone or in combination with Mamu-AG1*05:01, -AG2*01:02, and -B*45:03. Taken together with earlier studies defining similar ligands for KIR3DL05*008 (71, 73–75), at least seven rhesus macaque KIRs are known to encode receptors for Mamu-A or -AG ligands (Table I). We also identified Mamu-A1*012:01 as a ligand for KIR3DLw03*002 and KIR3DL02*004, which belong to a distinct group of receptors with a characteristic deletion of 3 aa in the D0 domain (Table I).
All of the KIRs that interact with Bw4 ligands recognize Mamu-B*041:01 and -B*065:01 with varying efficiency, suggesting that these molecules may provide a common reference for this group of receptors (Table I). However, interactions with other Mamu-Bw4 molecules were more variable. Several KIRs, including KIR3DL11*012 and KIR3DS04*003, lack detectable responses to Mamu-B*007:01, -B*022:01, or -B*043:01. Differences in the magnitude of KIR-CD3ζ JNL cell responses and KIR-Fc staining also suggest variation in the strength of these interactions. Although most of the KIRs responding to Mamu-Bw4 ligands are inhibitory, interactions with a couple of activating KIRs (KIR3DS04*003 and KIR3DSw39*002) were also observed (Table I). Thus, differences in the preferential recognition of individual Mamu-Bw4 molecules, variation in the strength of these interactions, and the identification of inhibitory and activating KIRs that respond to this group of molecules indicate extensive functional diversification of this group of receptors.
Rhesus macaque MHC class I haplotypes typically express 2 or 3 Mamu-A genes and 4–11 Mamu-B genes that can be classified as major or minor loci based on their transcriptional abundance (30–32). The Mamu-Bw4 molecules identified as KIR ligands are products of transcriptionally abundant genes (32). However, phylogenetic and segregation analyses suggest that they are distinct from the Mamu-B genes that encode molecules known to present viral peptides to CD8+ T cells (30–33, 84). This is further supported by unique polymorphisms in the cytoplasmic domains of Mamu-B*007:01, -B*022:01, -B*041:01, -B*043:01, and -B*065:01 that are not present in the cytoplasmic tails of molecules such as Mamu-B*008 and -B*017 that are known to present SIV epitopes for recognition by CD8+ T cells (33, 84). We previously demonstrated that these cytoplasmic domain polymorphisms prevent MHC class I downmodulation by SIV Nef (85). Selective downmodulation of HLA-A and -B (but not HLA-C) by HIV-1 Nef reduces the susceptibility of virus-infected cells to CD8+ T cells (6, 7, 86). Although the Vpu proteins of certain HIV-1 isolates may downmodulate HLA-C at later stages of infection (87, 88), HLA-C is initially retained on the cell surface, presumably to maintain interactions with inhibitory KIR (6). In a similar manner, SIV Nef downmodulates Mamu-A and -B molecules known to present viral peptides to CD8+ T cells, but not the products of Mamu-Bw4 genes that serve as ligands for multiple KIRs (74, 85). Hence, parallels in both immunogenetics and virology suggest functional specialization, whereby the products of certain Mamu-B genes play a dominant role in restricting CD8+ T cell responses whereas others serve as KIR ligands to modulate NK cell responses.
Another group of rhesus macaque KIRs interacts with Mamu-A–related ligands, including certain Mamu-AG allotypes and in some cases Mamu-B*045:03. KIR3DL07*004, KIR3DS01*003, KIR3DS02*004, KIR3DS06*002, KIR3DSw07*001, and KIR3DSw09*003 all recognize Mamu-AG2*01:01 and -AG6*04:01 (Table I). Additional interactions with Mamu-AG1*05:01 were detectable for KIR3DS02*004, KIR3DS06*002, and KIR3DSw09*003 and with Mamu-AG2*01:02 for KIR3DL07*004, KIR3DS02*004, and KIR3DSw09*003 (Table I). Mamu-B*045:03 was also identified as a ligand for KIR3DL07*004, KIR3DS02*004, and KIR3DSw09*003 (Table I). Although Mamu-B*045:03 is a product of a Mamu-B allele, a region of its α1 domain appears to be derived from an MHC-A gene. Thus, the hybrid origins of Mamu-B*045:03 probably account for its recognition by these KIRs.
In contrast to KIRs with specificity for Mamu-Bw4, most receptors for Mamu-AG are activating. Five of the seven rhesus macaque KIRs that interact with Mamu-AG2*01:01 and -AG6*04:01 are products of KIR3DS genes (Table I). In humans, the ligation of activating lineage III KIRs (KIR2DS) and KIR2DL4 on maternal NK cells of the placental decidua by HLA-C and soluble HLA-G stimulates the release of proinflammatory and proangiogenic factors that promote placental vascularization (20, 21, 45). Because macaques do not express orthologs of HLA-C or -G (28, 29, 38), it is conceivable that KIR3DS interactions with Mamu-AG play a similar role in placental development. Unlike human KIRs, which can be broadly categorized into inhibitory or activating haplotypes based on fixed differences in the number of activating genes, KIR haplotypes in macaques are much more variable (82). There are typically 4–17 expressed KIR genes per haplotype in the rhesus macaque, including at least one activating gene (56, 82). The expansion of lineage II KIRs that encode receptors for Mamu-AG may therefore ensure that every animal has at least one activating KIR capable of engaging this nonclassical molecule in the placenta.
The identification of Mamu-A1*012:01 as a ligand for KIR3DLw03*002 and KIR3DL02*004 provides at least one MHC class I ligand for a phylogenetically distinct group of receptors with a characteristic deletion in D0. Although Mamu-A1*012:01 was the only ligand identified for these KIRs among the molecules tested in this study, other ligands may be discovered by screening additional rhesus macaque MHC class I molecules. Comparison of the D0 domains of KIRs expressed in other primate species revealed that residues 37–39 are also absent from the lineage II KIRs of apes and humans. This suggests that hominid lineage II KIRs evolved from a KIR gene with a corresponding 9-bp deletion in exon 3 that was present in the common ancestor of apes and Old World monkeys. Further inspection indicates that this deletion results in polymorphic differences in potential N-linked glycosylation (PNG) sites that are close to a predicted MHC class I contact site (89). Whereas most rhesus macaque KIRs have two tandem PNG sites (N36FTN39FT), KIR3DL02, KIR3DLw03, KIR3DLw34, and KIR3DS05 only contain a single PNG site in this region (N36FT) (Fig. 1). Furthermore, because of an additional threonine-to-methionine substitution at residue 38 (N36FM), human lineage II KIRs (KIR3DL1 and KIR3DL2) do not have a PNG site in this region. Hence, these species-specific differences in glycosylation may influence KIR–MHC class I interactions.
The present study more than doubles the number of rhesus macaque KIRs with defined MHC class I ligands. At least one MHC class I ligand has now been identified for KIRs representing 16 different lineage II KIR genes in animals of Indian origin. These findings reveal a remarkable expansion of lineage II KIRs with broad specificity for Mamu-Bw4 or -AG ligands and identifies previously uncharacterized interactions with Mamu-AG, -B*045, and -A1*012 allotypes. The recognition of overlapping but nonredundant sets of ligands by both inhibitory and activating KIRs indicates extensive diversification of these receptors in concert with an expanded complement of Mamu-A and -B genes. These observations advance our basic understanding of KIR and MHC class I coevolution in primates and provide a valuable foundation for studying NK cell responses in the rhesus macaque as a preclinical model.
Disclosures
The authors have no financial conflicts of interest.
Footnotes
This work was supported by the U.S. Public Health Service grants AI095098, AI155163, AI161816, AI121135, AI148379, and AI098485 (to D.T.E.). Additional support was provided by U.S Public Health Service Grant OD011106 to the Wisconsin National Primate Research Center. D.T.E. is an Elizabeth Glaser Scientist of the Elizabeth Glaser Pediatric AIDS Foundation and a University of Wisconsin Medical Foundation Professor of Pathology and Laboratory Medicine.
The online version of this article contains supplemental material.